This research paper describes the creation and evaluation of the Multimodal Image Dataset for AI-Based Skin Cancer (MIDAS), a significant new resource for the development and testing of artificial intelligence models in dermatology. MIDAS is notable for being the largest publicly available dataset of biopsy-proven skin lesions that includes paired dermoscopic and clinical images, reflecting a real-world clinical workflow. The study evaluates the performance of several existing AI models using MIDAS, finding that their performance often decreases on this more diverse and prospectively collected data compared to previous benchmarks. The authors emphasize the importance of rigorous external validation of AI models in medical settings before widespread use and highlight MIDAS’s potential to improve the generalizability and clinical applicability of future dermatology AI tools.
LINK : https://ai.nejm.org/doi/full/10.1056/AIdbp2400732
Q: What is the MIDAS dataset? A: MIDAS stands for the Melanoma Research Alliance Multimodal Image Dataset for AI-based Skin Cancer. It is a multimodal image dataset designed for benchmarking artificial intelligence (AI) models used in skin cancer detection and diagnosis.
Q: Why was the MIDAS dataset created? A: The MIDAS dataset was created to address the need for high-quality, diverse datasets for developing and testing AI algorithms in dermatology. Most existing dermatology AI models have been built on proprietary, siloed data, often from a single site with only one image type (either clinical or dermoscopic). This contrasts with dermatologists’ real-world workflow, which is multimodal, combining clinical examinations, dermoscopy, and patient data. The lack of rigorous external benchmark evaluations and access to transparent, well-annotated multimodal datasets has limited the adoption and validation of multimodal AI strategies, hindering their clinical utility and the ability to uncover algorithmic biases.
Q: What kind of data does MIDAS contain? A: MIDAS is a multimodal dataset containing paired dermoscopic and clinical images of biopsy-proven skin lesions. Clinical images were acquired at both 15-cm and 30-cm distances. The dataset also includes well-annotated, patient-level clinical metadata and histopathologic confirmation for biopsied lesions. Clinical metadata includes information such as sex at birth, age, Fitzpatrick skin type, personal history of melanoma, anatomic location, and the lesion’s length and width. It also includes unbiopsied control lesions with favored diagnoses agreed upon by independent dermatologists.
Q: How was the MIDAS dataset built? A: MIDAS was built through a prospective recruitment process at Stanford University and the Cleveland Clinic Foundation between August 18, 2020, and April 17, 2023. Patients with skin lesions requiring a biopsy or previously identified as concerning were eligible and provided written informed consent. Standardized digital photography, including clinical images at 15-cm and 30-cm distances and dermoscopic images using specific devices, was performed. Reference labels for biopsied lesions were based on histopathologic diagnoses, reviewed by board-certified dermatopathologists. For unbiopsied lesions, a second independent board-certified dermatologist reviewed and agreed upon the managing physician’s favored diagnosis.
Q: What types of skin lesions are included in MIDAS? A: The images in MIDAS represent a diverse range of lesions typically seen in general dermatology clinics, including malignant, benign, and inflammatory types. Specific types included are melanocytic nevi (22.4%), invasive cutaneous melanomas (4.4%), and melanomas in situ (4.5%). The dataset also includes lesions like basal cell carcinoma (BCC), squamous cell carcinoma (SCC), squamous cell carcinoma in situ (SCCIS), actinic keratosis (AK), dermatofibroma, and seborrheic keratosis (SK). The dataset includes lesions with secondary changes like hemorrhagic crusts or excoriations, reflecting real-world clinical variability.
Q: How can MIDAS be used? A: MIDAS serves as a benchmark tool for external validation and comparative analysis of AI algorithms. Its multimodal and prospectively recruited nature makes it suitable for evaluating models that utilize different combinations of image types and clinical metadata. It allows researchers to assess the generalizability and real-world performance of models. MIDAS also facilitates the assessment of clinically relevant questions, such as the impact of image acquisition parameters like camera distance on algorithm performance.
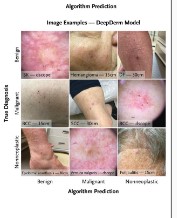
Q: What were the key findings when using MIDAS to evaluate AI models? A: An evaluation of four previously published state-of-the-art (SOTA) AI models on MIDAS revealed a decline in performance for all models compared with their original published metrics. Performance declines were particularly noted in sensitivity for malignancy and overall accuracy across three of the four algorithms evaluated. For example, ModelDerm’s AUROC dropped from 0.86 to 0.68, and DeepDerm’s mean accuracy dropped from 72.1% to 60.4%. Misclassification patterns showed that malignant lesions were often misclassified as nonneoplastic (inflammatory) by DeepDerm. Benchmarking also showed that AI model performance can be negatively impacted when classifiers trained on narrower disease distributions are challenged with a broader range of diagnoses found in MIDAS. Ensemble models using multiple modalities showed promising results in some metrics, like increased F1 scores and improved AUROC and specificity for a logistic regression metamodel.
Q: How did AI model performance compare to clinicians on MIDAS? A: As a baseline, dermatologists achieved 79% accuracy in identifying malignant lesions based on their top-one diagnosis using MIDAS cases. This accuracy increased to 91.1% when considering their top three diagnoses. On a challenging subset of lesions where all tested AI models incorrectly classified every image at top-one diagnosis, clinicians demonstrated 51.9% accuracy at top-one diagnosis. Dermoscopic images yielded higher sensitivity than clinical ones for clinicians.
Q: What are the strengths of the MIDAS dataset? A: Key strengths of MIDAS include that it is publicly available and prospectively recruited. It features systematically paired dermoscopic and clinical images, including clinical images at different distances. It provides well-annotated, patient-level clinical metadata and is supported by histopathologic confirmation for biopsied lesions. Being a dual-center dataset with diverse lesions seen in general dermatology, it mirrors real-world clinical scenarios more accurately than retrospective datasets. It includes lesions with secondary changes that reflect real-world variability. MIDAS serves as a standard for future multimodal benchmarks and facilitates the evaluation of models intended for broad diagnostic settings. Its creation followed best practices for image acquisition, expert data labeling (including in-person skin tone labeling), and ethical considerations.
Q: What are the limitations of the MIDAS dataset? A: Current limitations of MIDAS include a lower representation of Fitzpatrick skin types IV through VI and an age-related sampling bias toward adult cases. The lower representation of skin types IV-VI is likely due to lower incidence of skin cancer in people of color and disparate access to care at academic medical centers. The dataset also lacked paired dermatopathological data, which would have further enhanced its multimodality.
Q: Who funded the creation of MIDAS? A: The research supporting MIDAS was funded by the L’Oréal Dermatological Beauty Brands-Melanoma Research Alliance Team Science Award and other sources, including philanthropic funding from the David Mair and Vanessa Vu-Mair Artificial Intelligence in Skin Cancer Fund and the Tal and Cinthia Simon Melanoma Research Fund at Stanford Medicine. It also received a Stanford Human-Centered Artificial Intelligence Google Cloud Credits Grant and was supported with resources from the Veterans Affairs Palo Alto Health Care System.